ARAQUM – Arabidopsis RNA Quality Mechanisms
TEAM Programme
BACKGROUND
RNA surveillance, or quality control, represent mechanisms that are able to recognise and eliminate defective, potentially harmful, RNA molecules, mainly mRNAs but also non-coding RNAs. Up to date several types of RNA quality processes have been described, including Nonsense Mediated Decay (NMD) that degrades mRNAs with pre-mature termination codons (PTC), Non-Stop Decay (NSD) removing messages lacking the stop codon, No-Go decay (NGD) clearing out molecules with stalled ribosomes, and finally pathways operating in the nucleus to eliminate aberrant, superfluous or incorrectly processed RNAs. The impact of properly functioning quality control systems has now only started to emerge. It is estimated that around 30% of human alternatively spliced messages contain PTCs and in several cases such nonsense mRNAs lead to loss of function and contribute to diseases.
RNA quality control by NMD occurs in all eukaryotes, but factors involved, definition and recognition of PTC and mechanisms of RNA decay differ. In mammals, a splicing-dependent pathway predominates and an exon junction complex (EJC) plays a major role in recognition of the nonsense mRNAs by UPF proteins and translation termination factors that arrest translation and target transcripts for degradation. In contrast, in yeast or Drosophila discrimination between normal and PTC-containing mRNA relies on the correct interaction between the translation termination factors and proteins bound to the message 3′ UTR (so called “faux 3’UTR” model). mRNA substrates targeted for decay are usually degraded by two pathways with varying contribution in different organisms: 5’→3′ decay that involves removal of the cap structure by the decapping complex DCP1/DCP2 followed by digestion by XRN1; or 3’→5′ decay carried out by the exosome. In some cases, degradation is initiated by the endonucleolytic cleavage near the PTC and continued by the same exonucleases.
Surprisingly little is known about NMD in plants. A few major players, such as UPF1-3 proteins, SMG-7, Mago and Y14, have been identified and the PTC recognition rules by both intron-dependent and long 3’UTR modes have been described, but the majority of questions are still unanswered. This includes characterisation of decay pathways, contribution of the EJC and other components, and finally, location of the process within the cell. As plants differ noticeably from other organisms (sessile behaviour, development depending on external conditions, dormancy programmes etc.), it is likely that also RNA surveillance pathways will exhibit some distinct features. There are indications that plant NMD may involve the nucleolar phase via nucleolus-associated sub-organelles called Cajal Bodies (CB) and be somehow linked to RNA silencing.
OBJECTIVES and OUTCOMES
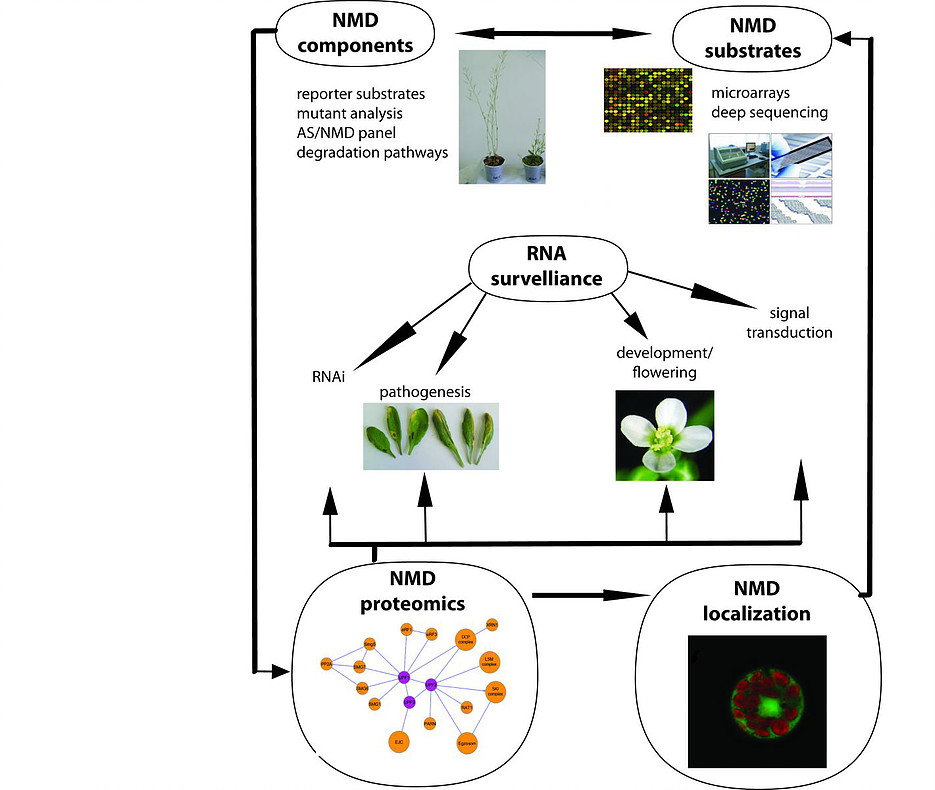
Major aim of the project is to dissect the mechanisms of mRNA decay in a model plant organism Arabidopsis thaliana during RNA surveillance pathways, with the focus on the major mRNA quality mechanism Nonsense Mediated Decay.
This will be achieved by the analysis of both endogenous and reporter nonsense containing substrates in Arabidopsis mutants in known or putative RNA metabolic enzymes and factors to identify those with direct roles in NMD and other surveillance processes. We will identify new or auxiliary elements of this machinery by purifying interacting components using tagged proteins. Finally, we will participate in studying the effects of defective NMD on plant development and hormone signalling and in establishing the correlations between NMD and RNA interference (RNAi).
The following aspects will be addressed: (1) Identification of RNA factors involved in plant NMD using a large collection of RNA metabolic mutants and testing endogeonous and reporter substrates as well as via the Alternative Splicing/NMD panel (AS/NMD); (2) Establishing a proteome of the NMD network by purification of interacting components using tandem affinity and MS methods; (3) Correlation of NMD with other RNA pathways, including RNAi; (4) Determining links between NMD and plant development, signaling and disease; (5) Analysis of NMD subcellular localisation and dynamics.
This projects will generate a comprehensive description of RNA decay in plant NMD, including the role and cellular location of enzymes and cofactors active in NMD, links with other surveillance pathways, rules of substrate recognition, significance of NMD in plant development, hormone signalling and pathogen resistance, as well as networking between various RNA-related processes in the control of gene expression in plants.
INTERNATIONAL COOPERATION
John Brown, University of Dundee (at Scottish Crop Research Institute), Dundee, UK
Brendan Davies, University of Leeds, Leeds, UK
Pamela Green, Delaware Biotechnology Institute, Newark, USA
Jacek Hennig, Magdalena Krzymowska, Institute of Biochemistry and Biophysics Polish Academy of Science, Warsaw, Poland
Peter Shaw, John Innes Centre, Norwich, UK
Daniel Silhavy, Agricultural Biotechnology Center, Gödöllõ, Hungary
Gordon Simpson, University of Dundee, Dundee, UK
Andrzej Wierzbicki, University of Michigan, Ann Arbor, USA
TEAM MEMBERS:
Group leader
Joanna Kufel, PhD
Postdocs
Izabela Wawer, PhD
Monika Zakrzewska-Płaczek, PhD
PhD students
Michał Krzysztoń, MSc
Paweł Sikorski, MScEng
Elżbieta Turek, MSc
Aleksandra Kwaśnik, MSc
MSc students
Dorota Kawa, BEng
Emilia Niemiec, BEng
Stanisław Łoboziak, BSc
Hanna Ollik, BSc
Zofia Janicka, BSc
Foundation for Polish Science Team Programme co-financed by the EU Innovative Economy Operational Programme 2007-2013.