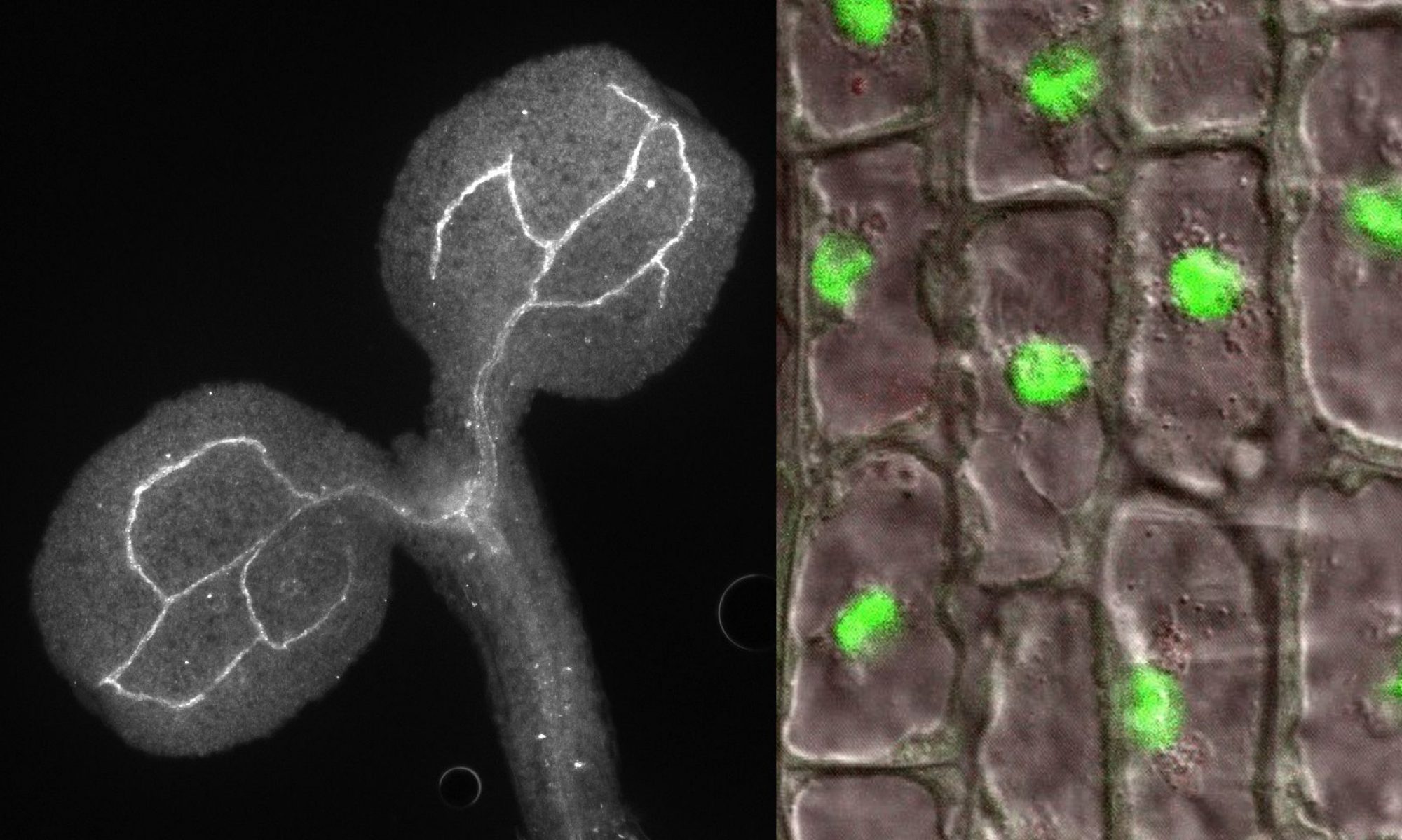
As semi-autonomous organelles mitochondria have their own DNA and translational machinery, but mitochondrial DNA encodes only for a few polypeptides, while the majority of mitochondrial proteins are nuclear-encoded, translated in the cytosol and imported into mitochondria. The best-characterized mechanism of mitochondrial targeting involves specialized translocases, which deliver proteins containing the N-terminal mitochondrial targeting signal (MTS) as unfolded precursors. Many mitochondrial proteins do not contain an N-terminal presequence with the MTS, but may carry unidentified or ill-defined internal targeting sequences and the mechanisms of their translocation to mitochondria remain elusive.
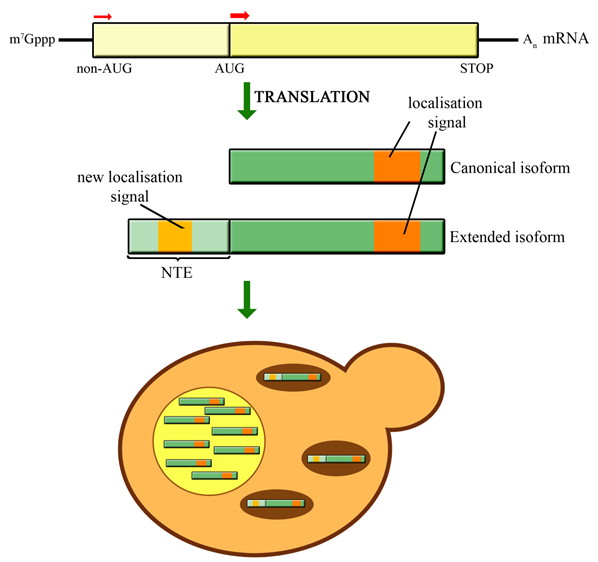
Utilization of non-AUG alternative translation start sites is most common in bacteria and viruses, but it has been also reported in other organisms. This phenomenon increases protein complexity by allowing expression of multiple protein isoforms from a single gene. Using bioinformatics tools, we provide evidence that non-AUG translation initiation is much more prevalent than previously anticipated and may apply to as many as few thousands of proteins. Several hundreds of candidates are predicted to gain MTS, generating a hidden cache of mitochondrial proteins. We confirmed mitochondrial localization of a number of proteins previously not recognized as mitochondrial whose standard forms do not contain an MTS. Our data highlight the potential of non-canonical translation initiation in expanding the capacity of the mitochondrial proteome and probably also other cellular features.
Now we focus on several RNA-related factors and complexes with potential mitochondial localization and function. These incluse LSM proteins, TRAMP components and ribosome biogenesis factors.
Publications from this project
Utilization of non-AUG alternative translation start sites is most common in bacteria and viruses, but it has been also reported in other organisms. This phenomenon increases proteome complexity by allowing expression of multiple protein isoforms from a single gene. In Saccharomyces cerevisiae, a few described cases concern proteins that are translated from upstream near-cognate start codons as N-terminally extended variants that localize to mitochondria. Using bioinformatics tools, we provide compelling evidence that in yeast the potential for producing alternative protein isoforms by non-AUG translation initiation is much more prevalent than previously anticipated and may apply to as many as a few thousand proteins. Several hundreds of candidates are predicted to gain a mitochondrial targeting signal (MTS), generating an unrecognized pool of mitochondrial proteins. We confirmed mitochondrial localization of a subset of proteins previously not identified as mitochondrial, whose standard forms do not carry an MTS. Our data highlight the potential of non-canonical translation initiation in expanding the capacity of the mitochondrial proteome and possibly also other cellular features.