Project NCN 2014/13/B/NZ3/00405
Principal investigator: dr Anna Golisz
Research project objectives:
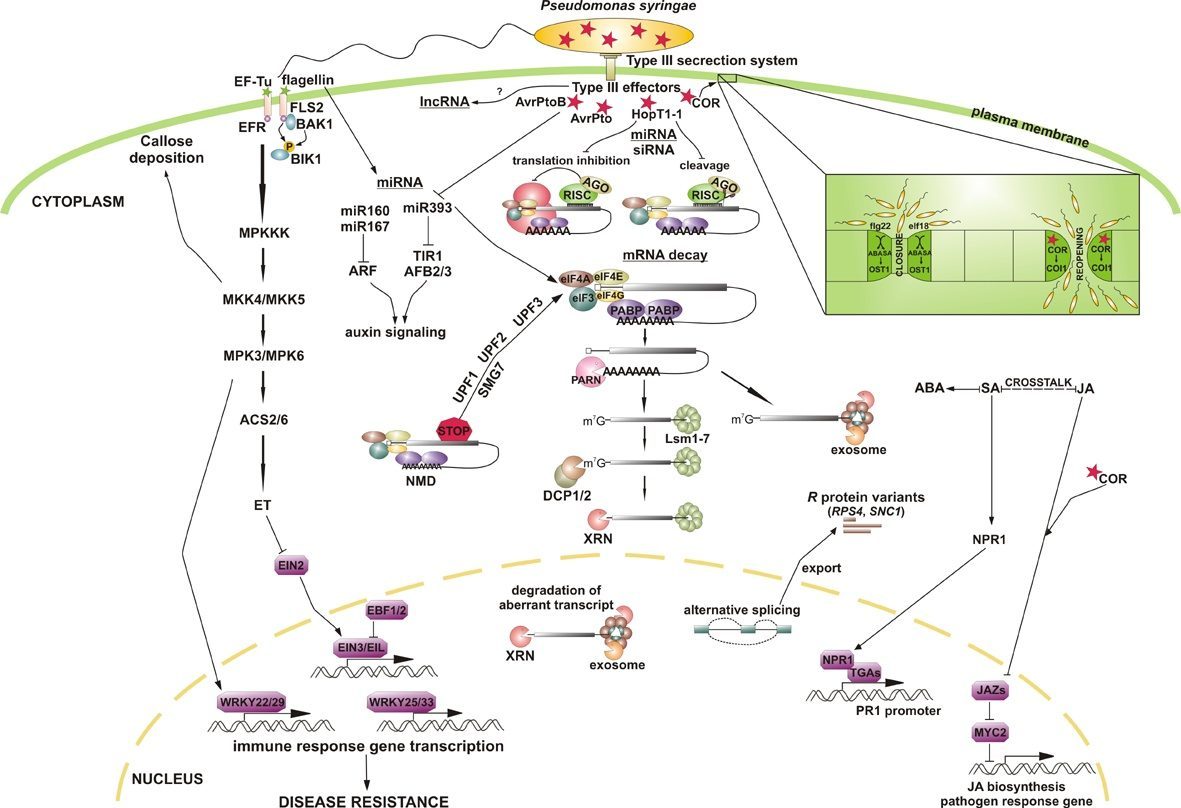
The proposed research involves the systematic and comprehensive analysis of Arabidopsis thaliana mutants in specific RNA metabolism pathways, including mRNA splicing and degradation, to identify defects that lead to changes in the response to biotic stress induced by the infection of Pseudomonas syringae pv. tomato DC3000. The proper cellular signaling in each organism depends on the optimal control of gene expression, not only under normal conditions but also under stress. Recent studies have shown that a number of mutants in major RNA quality control pathways, such as NMD, show defects in pathogen response, but the exact mechanism of this correlation is not clear. Also a comprehensive scheme of links between regulation of gene expression and activation of the immune response is lacking. Two important players in these pathways are the cascade of phosphorylation signaling via MAP kinases, and small RNAs, siRNA and miRNA, through silencing of gene expression (RNAi) on the transcriptional or posttranscriptional level. Furthermore, recently a number of long noncoding RNAs (lncRNAs) have been identified in plants and several of these may play an important role in antibacterial immunity. Therefore, we propose to identify a network of interdependencies and connections between RNA factors and key regulators of pathogenesis, such as MAP kinases or ncRNAs.
Research project methodology:
The tasks of the project will be implemented through the analysis of the sensitivity of selected Arabidopsis mutants with defects in RNA processing and degradation factors to infection by P. syringae. The resistance of investigated mutants will be tested by the rate of bacterial growth in infected leaves. In addition, by microscopic observations we will determine the relationship between the response of stomatal apertures, the process of callose deposition and sensitivity to pathogen. The next step will be to evaluate the level of plant hormones (ET, SA, JA, ABA), which are involved in regulating the response to the pathogen. Furthermore, we will assess the molecular response during biotic stress induced by different effectors by treating plants with flg22, elf18, COR and SA, which mimics the action of pathogen infection.
Mutants showing significant changes in sensitivity to pathogen infection will be analysed further by northern blot and quantitative RT-PCR for accompanying alterations in molecular phenotypes, that is the kinetics of activation, level and stability of known pathogenesis markers and pathogen-related WRKY transcription factors mRNAs. If abnormal response to biotic stress occurs in mutants defective in pre-mRNA splicing, particularly in alternative splicing, the level and pattern of R protein AS forms will be examined. These proteins play an important role in resistance to infection, and changes in their expression may give rise to irregularities in the response to pathogen. We also intend to determine whether defective response to pathogen in RNA metabolism mutants is associated with impaired MAP kinase cascade that regulates the progress of infection. For this purpose, in selected mutants we determine the level and stability of MAPK mRNAs (northern blot, qRT-PCR) and their kinase activity (in gel kinase assay, western blot), as well as changes in phosphorylation status of known and potential MAPK substrates in RNA metabolism factors. The level of phosphorylation of these factors will be determined in Arabidopsis protoplasts of wild-type and mutant plants by immunoprecipitation of GFP-tagged proteins and detection of phosphorylated residues using Pro-Q Diamond staining or targeted mass spectrometry.
As ncRNA play an increasingly recognized part in many cellular processes, we plant to assess their role in plant pathogen defense by carrying out high-throughput sequencing (RNA-Seq) of total and small RNA libraries following the treatment with P. syringae for selected Arabidopsis mutants with the most significant changes in sensitivity to pathogen infection. This approach will permit not only to evaluate the contribution of short RNAs, such as siRNAs or miRNAs, but also to identify lncRNAs with pathogenesis regulatory potential. RNA sequencing data will be confirmed by northern blots and RT-PCR, also with respect to the stability of pathogen-related mature and precursor ncRNAs. In turn, characterization of lncRNA target genes will help us to establish new components in plant immunity signaling pathways, which will form the basis for future research.
These integrated approaches will allow us to obtain a more complete picture of defense mechanisms occurring in plants and to ascertain their direct regulation by the processes of RNA metabolism.
Research project impact:
It is important for the development of civilization to understand how plants achieve immunity to infection by pathogenic organisms in order to effectively protect the world’s food production. Many international scientific groups investigate the mechanisms of plant responses to biotic stress, but despite several preliminary observations, a comprehensive analysis of the correlation between RNA metabolism and plant response to stress factors is still missing. Thus, the results of experiments planned in this project will allow us to demonstrate that RNA processing and degradation is an important factor in plant signaling during defense responses, thus contributing in a significant way to the knowledge in the field.