A subclass of yeast ncRNAs, namely small nuclear and nucleolar RNAs, are synthesized by RNA polymerase II as precursors, either from independent transcription units (sn/snoRNAs) or are released from polycistronic transcripts or introns in pre-mRNAs. All need to be accurately processed and assembled into RNP molecules. Another ncRNA types, including Cryptic Unstable Transcripts (CUTs), similarly to mRNAs often carry poli(A) tails but are usually rapidly degraded following synthesis. Both snoRNA accumulation and CUT decay occurs via pathway involving cleavage/termination giving way to trimming by the exosome, which is facilitated by the nuclear RNA surveillance complex TRAMP (Trf4/5+Air1/2+Mtr4). Termination is mediated by two machineries: Cleavage and Polyadenylation (CP), acting predominantly on coding genes, and Nrd1/Nab3/Sen1 (two RNA binding proteins and RNA helicase)- more specific for sn/snoRNAs.
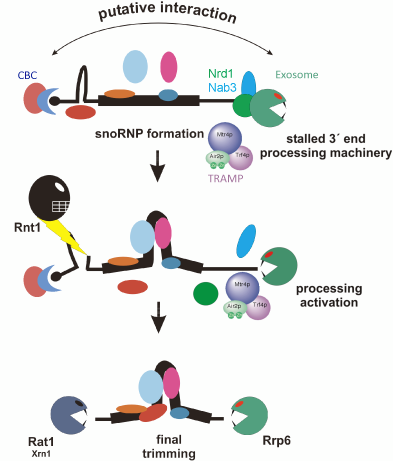
Our recent work on transcription termination and processing of snoRNAs revealed the mechanisms by which the same RNA polymerase II produces different RNA molecules, polyadenylated mRNAs and snoRNAs lacking poly(A) tails. We confirmed that termination of snoRNAs occurs predominantly at the specific Nrd1/Nab3-dependent site, while the second region acts as a fail-safe terminator. We showed that termination at both sites is followed by precursor polyadenylation by a major poly(A) polymerase Pap1 with the contribution by the alternative poly(A) polymerase Trf4. Another function of Tfr4 is to adenylate processing intermediates to facilitate their maturation by the exosome/Rrp6. We also discovered the important function of Trf4 and TRAMP independent of its polyadenylation activity, that is stabilization of the Nrd1/Nab3 association at snoRNA genes. This is a first demonstrated example of polyadenylation-independent activity performed by TRAMP.
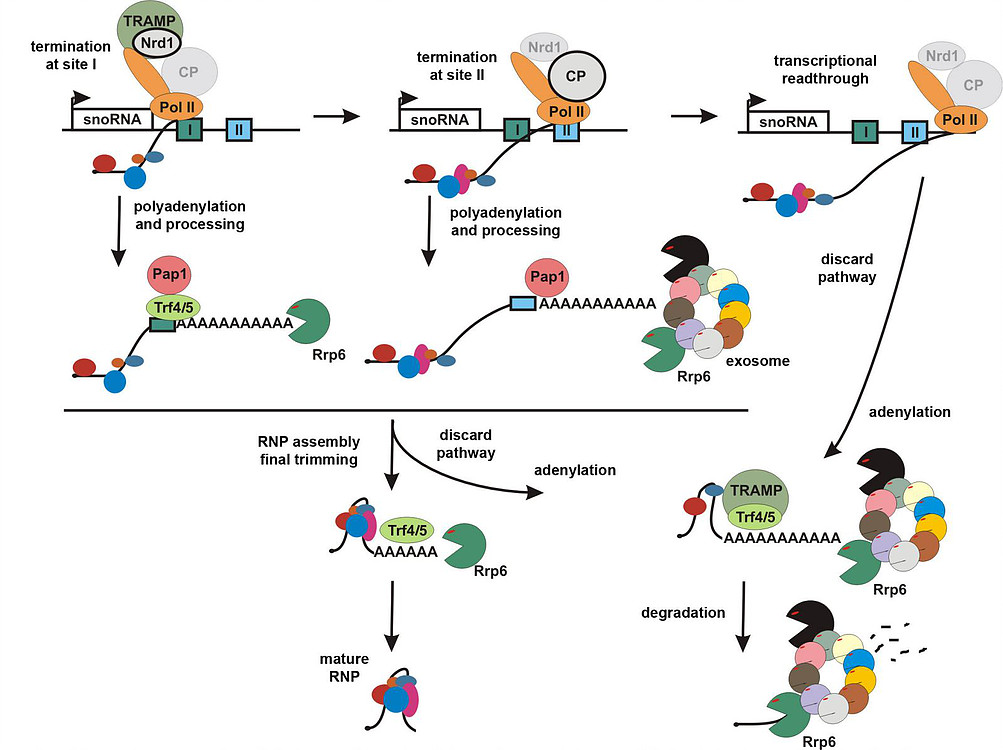
The major conclusion is that snoRNA precursors also become polyadenylated in a process linked to termination and the role of the poly(A) tails is to facilitate 3’ end processing by the nuclear exosome/Rrp6. 3’ end formation occurs via subsequent rounds of adenylations and trimmings until the progression of exonucleases is blocked by the accurately assembled RNP. In turn, defective molecules cannot stop the exosome and become degraded by the same machinery. This data unifies the process of transcription termination of Pol II transcripts and explains that poly(A) tails of intrinsically nonpolyadenylated RNAs are used for both processing and RNA quality control.
Most of boxC/D snoRNAs undergo 5′ processing through cleavage by Rnt1p and 5′-3′ trimming by exonucleases Rat1p and Xrn1p. Surprisingly, in the absence of Rnt1 or exonucleases 5′ unprocessed pre-snoRNAs also carry 3′ extensions composed of precursor sequences and oligo(A) tails. This suggests that processing of both snoRNA ends is coupled, most likely through cooperation of complexes interacting with their termini.