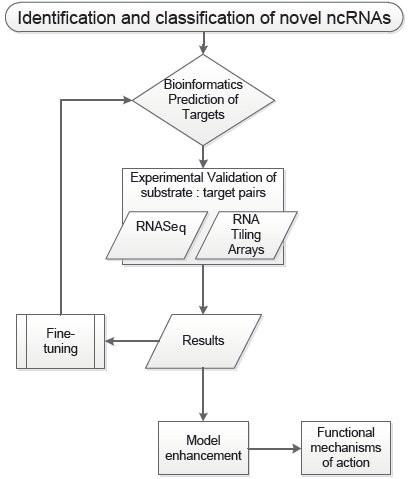
The project “Functional diversity of non-coding RNAs” realized by the NOCORE (Non-Coding RNA Enterprise) consortium aims at furthering our understanding of long non-coding RNAs (lncRNA) function in yeast and higher eukaryotic cells. Some of these ncRNAs are abundantly expressed in cancer cells and were proposed to have regulatory roles in signaling pathways. We want to unravel the full potential of natural lncRNAs, as well as those derived from housekeeping RNAs (hncRNAs) (rRNAs, tRNAs and other), in regulation of gene expression in Eukaryotes. This major objective encompasses the discovery of novel classes of ncRNAs, understanding the complexity of their synthesis and maturation as well as regulatory mechanisms, and finally deciphering their function by identifying the substrates they target and processes they act.
We propose to explore the universal occurrence, nature and function of eukaryotic lncRNAs and hncRNAs. The work is divided into 3 interdependent parts:
- Using global approaches to extend the repertoire of these ncRNAs. The analyses (tiling arrays and high-throughput sequencing) will be performed for cells deficient in specic RNA degradation pathways or in various physiological and stress conditions.
- Establishing the role and mechanisms of action of these ncRNAs. This will be addressed by searching for potential targets using bioinformatics tools and their experimental validation (functional analyses, localisation of novel ncRNAs) for the selected cases.
- In-depth analysis of the RNA-dependent gene cosuppression in yeast, which regulates the expression of mRNAs and rRNA. This will reveal how RNA silencing is achieved in a eukaryotic organism devoid of the RNAi machinery.