Mechanistic study of transcription regulation in Arabidopsis
Polish-Chinese Funding Initiative
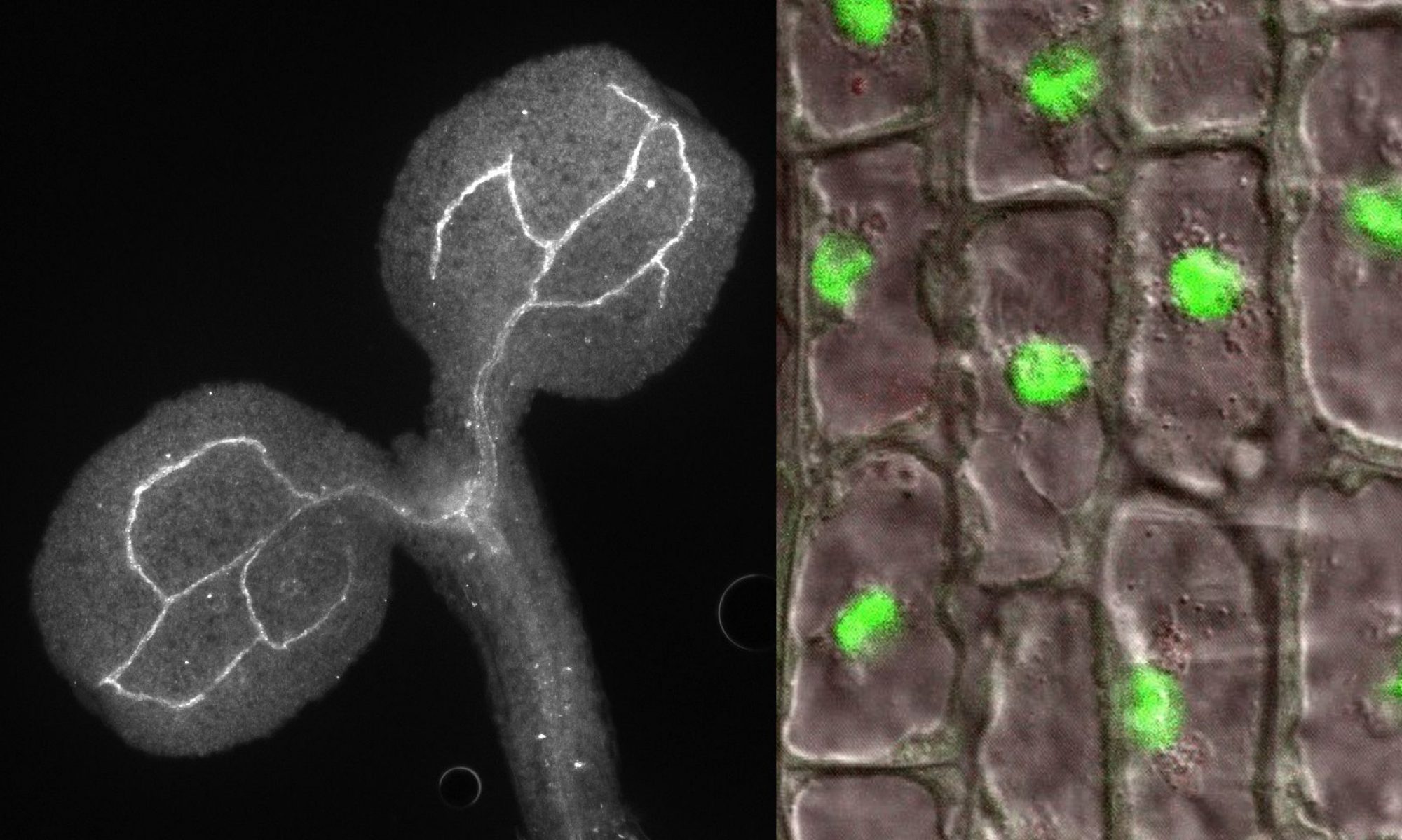
Coordinated and well-timed gene expression in all organisms requires precise regulation that occurs at different levels, including transcription and co-transcriptional processes. Although a few decades of research provided a deep insight into molecular bases of these mechanisms, there are new findings that add to a complex network of functional interactions. It became apparent that many enzymes and factors, including ribonucleases, RNA surveillance factors and auxiliary proteins, contribute to transcription regulation. In addition, it transpired recently that transcription machinery is organized in condensates generated via liquid-liquid phase separation (LLPS). The mechanism of condensate formation, their composition and exact role in gene expression regulation, particularly in plants, is still unclear. The proposed project involves comprehensive biochemical and functional studies of transcription regulation in Arabidopsis thaliana by three proteins, XRN3, DXO1 and RSA1, which were selected based on solid preliminary results obtained by both collaborating research groups. XRN3 5′-3′ exoribonuclease is mainly involved in the torpedo transcription termination mechanism of RNA polymerase II. DXO1 is an enzyme capable of removing the non-canonical NAD cap from RNAs, but its major functions are unrelated to catalytic activity. In turn, RSA1 is required for salt tolerance and forms liquid-like condensates that contain components of the transcription machinery and RNA-related proteins. Our previous studies and combined unpublished data clearly suggest that these proteins interact physically and/or functionally and are associated with Pol II activity. We envisage that they tightly cooperate in coupling different stages of the transcription cycle. The main objective of the proposal is to dissect the exact mechanism of the joint contribution of XRN3, DXO1 and RSA1 to transcription initiation, elongation and termination. In addition, based on their potential to undergo phase separation, we plan to explore the possibility that their function in transcription regulation is executed via condensate formation. Finally, we want to assess the impact of transcriptional activity of these factors on cellular processes and plant physiology, including response to environmental stresses.
DXO1 and non-canonical RNA cap structures
PROJECT IS FOUNDED BY THE POLISH NATIONAL SCIENCE CENTRE (NCN)
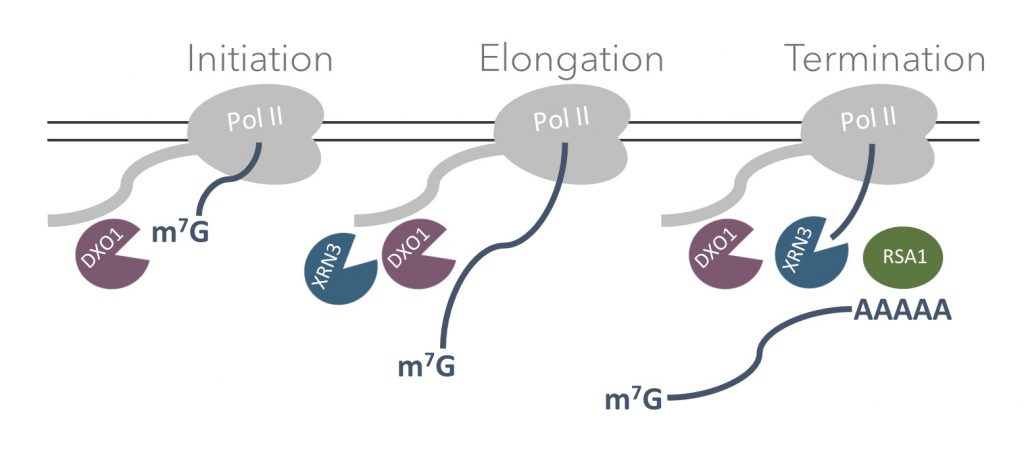
DXO/Rai1 proteins participate in mRNA 5′-end quality control (5’QC), removal of non-canonical NAD cap (deNADding) and maturation of fungal ribosomal RNA (rRNA). Our recent research described the biochemical activity and physiological significance of the Arabidopsis thaliana DXO1. Detailed enzymatic assays showed that DXO1 has strong deNADding and weak 5′-3′ exoribonuclease activities. Other biochemical properties characteristic of DXO/Rai1 homologs are inhibited in plants due to the presence of a single amino acid modification within the active site. DXO1 also contains a large, plant-specific N-terminal extension (NTE) that negatively affects its biochemical properties and participates in RNA binding. In addition, enzymatic activity of DXO1 is inhibited by adenosine 3′, 5′-bisphosphate (PAP), which is a component of chloroplast-to-nucleus retrograde signaling and an inhibitor of XRN 5′-3′ exoribonucleases.
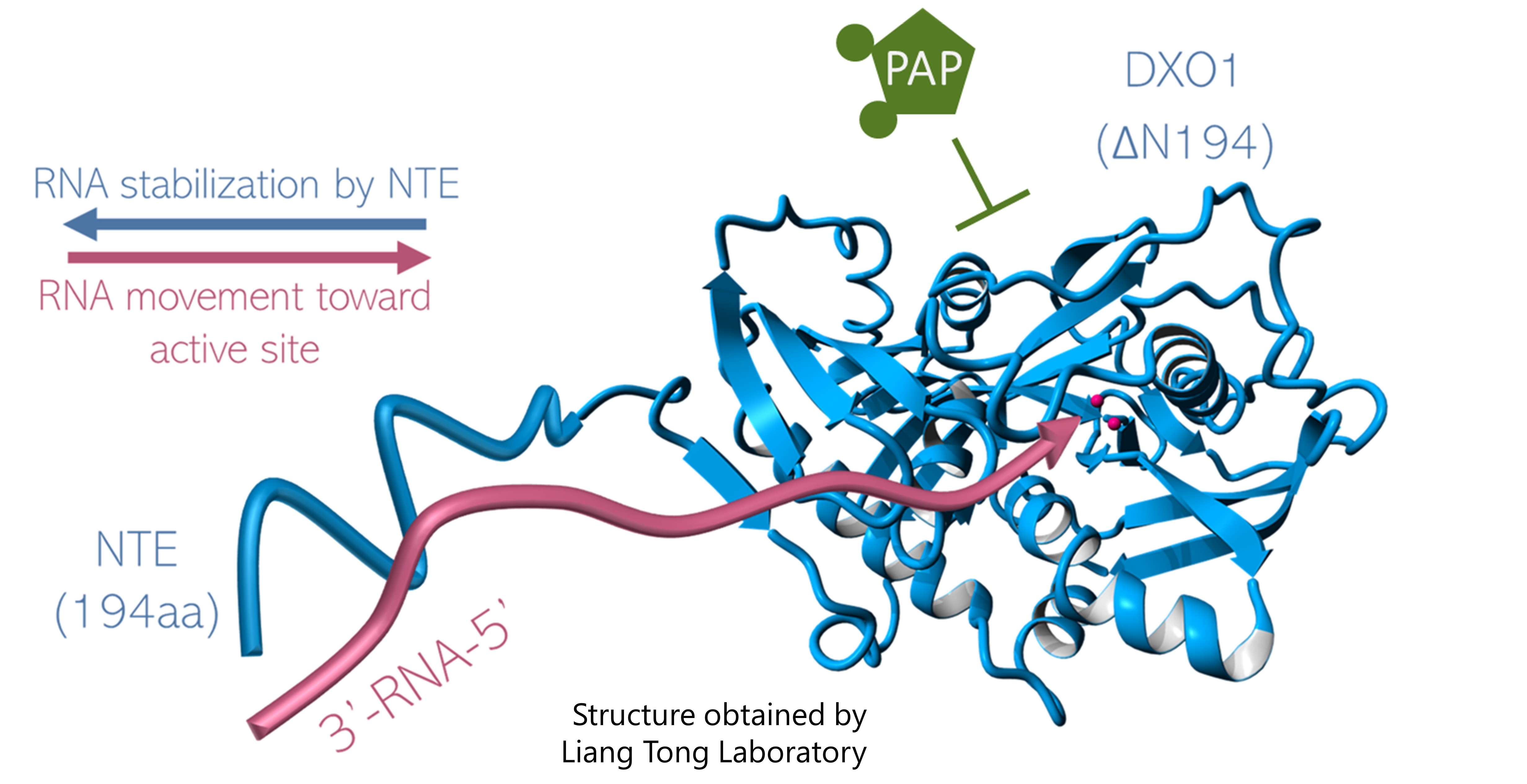
Currently, we work on the more detailed characterization of the function of DXO1, as well as other enzymes involved in the metabolism of non-canonical RNA caps.
Regulatory relationships between RNA metabolism and biotic stress defense in Arabidopsis thaliana
NCN 2014/13/B/NZ3/00405
It is important for the development of civilization to understand how plants achieve immunity to infection by pathogenic organisms in order to effectively protect the world’s food production. Our research involves the systematic and comprehensive analysis of Arabidopsis thaliana mutants in specific RNA metabolism pathways, including mRNA splicing and degradation, to identify defects that lead to changes in the response to biotic stress induced by the infection of Pseudomonas syringae pv. tomato DC3000.
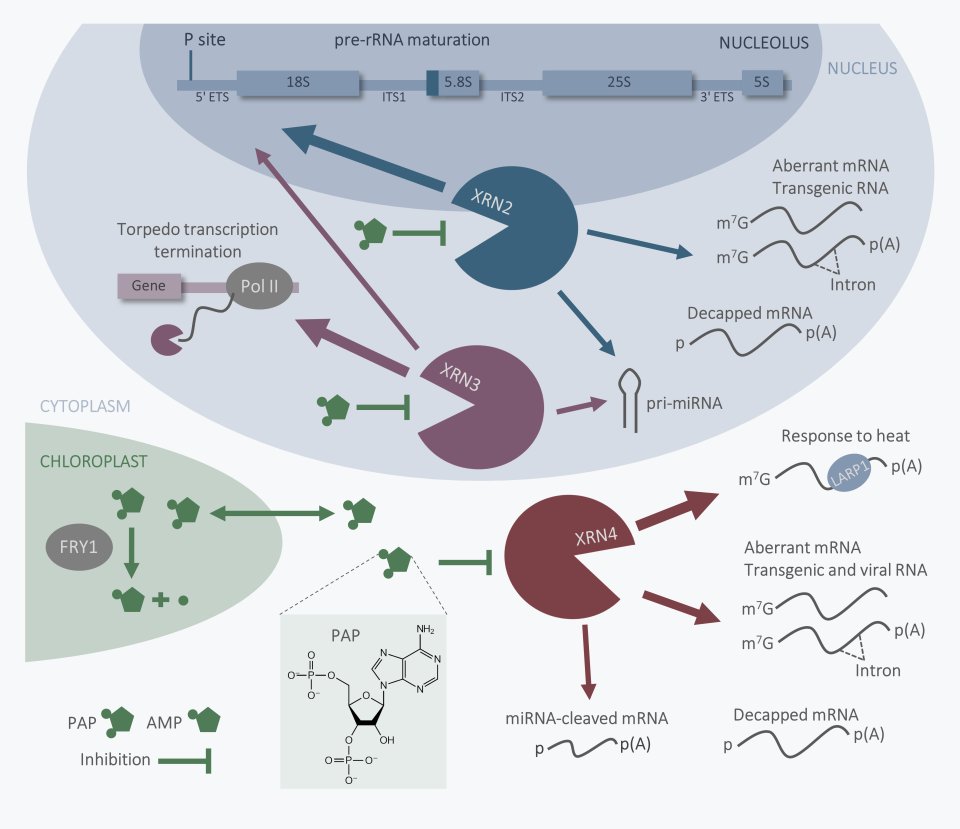
XRN family of 5'-3' exoribonucleases
These proteins are highly similar to each other but have distinct functions. We showed that AtXRN2, but not AtXRN3, has an unexpected role in pre-rRNA processing at the early endonucleolytic cleavage at site P that is carried out by an unknown component of a large U3-containing NF D ribonucleoprotein complex. Our data reveal that AtXRN2 performs initial shortening of the 5’ external pre-rRNA spacer prior to the cleavage, and this trimming is required to expose the site for processing. AtXRN2 also degrades polyadenylated rRNA precursors and excised pre-rRNA spacer fragments, contributing to the nuclear polyadenylation-dependent RNA surveillance pathway.
Nonsense-Mediated Decay (NMD) and splicing
One of the most important cytoplasmic RNA quality control pathway is Nonsense-Mediated mRNA Decay mechanism (NMD), which recognizes aberrant mRNAs carrying premature termination codons (PTC). Aberrant PTC-containing transcripts most often result from alternative splicing, mutations or transcription errors. The major function of NMD is to protect against accumulation of potentially harmful proteins, but it also regulates the expression of genetic information of normal transcripts and plays a role in response to stress. Therefore, NMD is not only an RNA quality mechanism, but also contributes to cellular homeostasis.